A bit more of Pandas¶
and some RegEx¶
SC 4125: Developing Data Products
Module-2: Basics of Pandas structures and manipulation
by Anwitaman DATTA
School of Computer Science and Engineering, NTU Singapore.
Disclaimer/Caveat emptor¶
- Non-systematic and non-exhaustive review
- Example solutions are not necessarily the most efficient or elegant, let alone unique
Positioning this module in the big picture¶
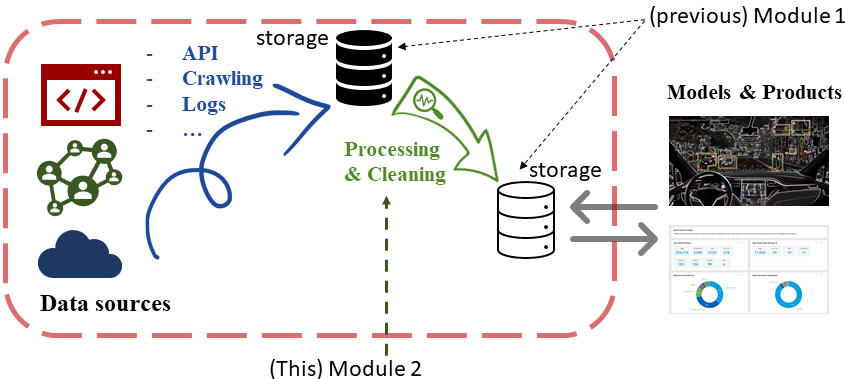

- Some basic tools for cleaning and manipulating your data: Apply, Lambdas, Merge, Groupby, Missing data, etc.
# housekeeping - imports necessary libraries
import numpy as np
import pandas as pd
import re
import datetime
Load data and carry out some preliminary cleaning¶
Let's load the same gapminder data we had worked with in the previous module, but from a .tsv file this time. Likewise, let's load the CountriesOfTheWorld data from a .csv file.
# load data
gapminderdatapath ='data/gapminder/' # change this to adjust relative path
gap_df = pd.read_csv(gapminderdatapath+'gapminder.tsv', sep='\t')
gap_df.sample(5)
| country | continent | year | lifeExp | pop | gdpPercap | |
|---|---|---|---|---|---|---|
| 761 | Israel | Asia | 1977 | 73.060 | 3495918 | 13306.619210 |
| 1263 | Reunion | Africa | 1967 | 60.542 | 414024 | 4021.175739 |
| 787 | Jamaica | Americas | 1987 | 71.770 | 2326606 | 6351.237495 |
| 868 | Lebanon | Asia | 1972 | 65.421 | 2680018 | 7486.384341 |
| 1473 | Sweden | Europe | 1997 | 79.390 | 8897619 | 25266.594990 |
# load data
countriesdatapath ='data/'
CoTW_df = pd.read_csv(countriesdatapath+'CountriesOfTheWorld.csv')
CoTW_df.sample(5)
| Country | Region | Population | Area (sq. mi.) | Pop. Density (per sq. mi.) | Coastline (coast/area ratio) | Net migration | Infant mortality (per 1000 births) | GDP ($ per capita) | Literacy (%) | Phones (per 1000) | Arable (%) | Crops (%) | Other (%) | Climate | Birthrate | Deathrate | Agriculture | Industry | Service | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 127 | Maldives | ASIA (EX. NEAR EAST) | 359008 | 300 | 1196,7 | 214,67 | 0 | 56,52 | 3900.0 | 97,2 | 90,0 | 13,33 | 16,67 | 70 | 2 | 34,81 | 7,06 | 0,2 | 0,18 | 0,62 |
| 10 | Aruba | LATIN AMER. & CARIB | 71891 | 193 | 372,5 | 35,49 | 0 | 5,89 | 28000.0 | 97,0 | 516,1 | 10,53 | 0 | 89,47 | 2 | 11,03 | 6,68 | 0,004 | 0,333 | 0,663 |
| 130 | Marshall Islands | OCEANIA | 60422 | 11854 | 5,1 | 3,12 | -6,04 | 29,45 | 1600.0 | 93,7 | 91,2 | 16,67 | 38,89 | 44,44 | 2 | 33,05 | 4,78 | 0,317 | 0,149 | 0,534 |
| 163 | Poland | EASTERN EUROPE | 38536869 | 312685 | 123,3 | 0,16 | -0,49 | 8,51 | 11100.0 | 99,8 | 306,3 | 45,91 | 1,12 | 52,97 | 3 | 9,85 | 9,89 | 0,05 | 0,311 | 0,64 |
| 185 | Slovakia | EASTERN EUROPE | 5439448 | 48845 | 111,4 | 0,00 | 0,3 | 7,41 | 13300.0 | NaN | 220,1 | 30,16 | 2,62 | 67,22 | 3 | 10,65 | 9,45 | 0,035 | 0,294 | 0,672 |
What is the list of countries in the gapminder and CoTW datasets?
gap_country_names_series=gap_df['country'].drop_duplicates()
print(gap_country_names_series.to_list()[:10]) #if you print the length, you will get a result of 142
['Afghanistan', 'Albania', 'Algeria', 'Angola', 'Argentina', 'Australia', 'Austria', 'Bahrain', 'Bangladesh', 'Belgium']
CoTW_country_names=CoTW_df['Country'].unique()
print(CoTW_country_names[:10]) #if you print the length, you will get a result of 227
['Afghanistan ' 'Albania ' 'Algeria ' 'American Samoa ' 'Andorra ' 'Angola ' 'Anguilla ' 'Antigua & Barbuda ' 'Argentina ' 'Armenia ']
# Let's get rid of the trailing space in the records
CoTW_df['Country']=CoTW_df.Country.apply(lambda x: x[:-1])
# Alternative: CoTW_df['Country']=CoTW_df.Country.apply(lambda x: x.rstrip())
# Alternate way would be to `strip' the space (in this case, rstrip would also work)
CoTW_country_names=CoTW_df['Country'].unique()
print(CoTW_country_names[:10])
['Afghanistan' 'Albania' 'Algeria' 'American Samoa' 'Andorra' 'Angola' 'Anguilla' 'Antigua & Barbuda' 'Argentina' 'Armenia']
Apply & Lambda¶
- We just saw the use of two very interesting tools (which often go hand-in-hand when processing a data frame):
- Apply: Apply a function along an axis of the data frame. It can also be used over a column.
- See details at the doc page
- Lambda: A small anonymous function. You can of-course use a named function inside your lambda, if you need something more complicated.
- Apply: Apply a function along an axis of the data frame. It can also be used over a column.
# load data
olymp_df=pd.read_html(r'https://en.wikipedia.org/wiki/2016_Summer_Olympics_medal_table')
olymp2016medals=olymp_df[2][:-1]
olymp2016medals.sample(5)
| Rank | NOC | Gold | Silver | Bronze | Total | |
|---|---|---|---|---|---|---|
| 84 | 78 | Trinidad and Tobago | 0 | 0 | 1 | 1 |
| 15 | 16 | Jamaica | 6 | 3 | 2 | 11 |
| 35 | 35 | Thailand | 2 | 2 | 2 | 6 |
| 0 | 1 | United States | 46 | 37 | 38 | 121 |
| 10 | 11 | Netherlands | 8 | 7 | 4 | 19 |
print(sorted(set(olymp2016medals['NOC'])-set(CoTW_country_names)) )
print(sorted(set(CoTW_country_names)-set(olymp2016medals['NOC'])))
# Note that several of the same countries are named or spelled differently in the two lists
['Bahamas', 'Brazil*', 'Chinese Taipei', 'Great Britain', 'Independent Olympic Athletes', 'Ivory Coast', 'Kosovo', 'North Korea', 'South Korea', 'Trinidad and Tobago'] ['Afghanistan', 'Albania', 'American Samoa', 'Andorra', 'Angola', 'Anguilla', 'Antigua & Barbuda', 'Aruba', 'Bahamas, The', 'Bangladesh', 'Barbados', 'Belize', 'Benin', 'Bermuda', 'Bhutan', 'Bolivia', 'Bosnia & Herzegovina', 'Botswana', 'Brazil', 'British Virgin Is.', 'Brunei', 'Burkina Faso', 'Burma', 'Cambodia', 'Cameroon', 'Cape Verde', 'Cayman Islands', 'Central African Rep.', 'Chad', 'Chile', 'Comoros', 'Congo, Dem. Rep.', 'Congo, Repub. of the', 'Cook Islands', 'Costa Rica', "Cote d'Ivoire", 'Cyprus', 'Djibouti', 'Dominica', 'East Timor', 'Ecuador', 'El Salvador', 'Equatorial Guinea', 'Eritrea', 'Faroe Islands', 'French Guiana', 'French Polynesia', 'Gabon', 'Gambia, The', 'Gaza Strip', 'Ghana', 'Gibraltar', 'Greenland', 'Guadeloupe', 'Guam', 'Guatemala', 'Guernsey', 'Guinea', 'Guinea-Bissau', 'Guyana', 'Haiti', 'Honduras', 'Hong Kong', 'Iceland', 'Iraq', 'Isle of Man', 'Jersey', 'Kiribati', 'Korea, North', 'Korea, South', 'Kuwait', 'Kyrgyzstan', 'Laos', 'Latvia', 'Lebanon', 'Lesotho', 'Liberia', 'Libya', 'Liechtenstein', 'Luxembourg', 'Macau', 'Macedonia', 'Madagascar', 'Malawi', 'Maldives', 'Mali', 'Malta', 'Marshall Islands', 'Martinique', 'Mauritania', 'Mauritius', 'Mayotte', 'Micronesia, Fed. St.', 'Moldova', 'Monaco', 'Montserrat', 'Mozambique', 'N. Mariana Islands', 'Namibia', 'Nauru', 'Nepal', 'Netherlands Antilles', 'New Caledonia', 'Nicaragua', 'Oman', 'Pakistan', 'Palau', 'Panama', 'Papua New Guinea', 'Paraguay', 'Peru', 'Reunion', 'Rwanda', 'Saint Helena', 'Saint Kitts & Nevis', 'Saint Lucia', 'Saint Vincent and the Grenadines', 'Samoa', 'San Marino', 'Sao Tome & Principe', 'Saudi Arabia', 'Senegal', 'Seychelles', 'Sierra Leone', 'Solomon Islands', 'Somalia', 'Sri Lanka', 'St Pierre & Miquelon', 'Sudan', 'Suriname', 'Swaziland', 'Syria', 'Taiwan', 'Tanzania', 'Togo', 'Tonga', 'Trinidad & Tobago', 'Turkmenistan', 'Turks & Caicos Is', 'Tuvalu', 'Uganda', 'United Kingdom', 'Uruguay', 'Vanuatu', 'Virgin Islands', 'Wallis and Futuna', 'West Bank', 'Western Sahara', 'Yemen', 'Zambia', 'Zimbabwe']
CoTW_df.loc[(CoTW_df.Country=="Bahamas, The"),'Country']='Bahamas'
CoTW_df.loc[(CoTW_df.Country=='Taiwan'),'Country']='Chinese Taipei'
CoTW_df.loc[(CoTW_df.Country=='United Kingdom'),'Country']='Great Britain'
CoTW_df.loc[(CoTW_df.Country=="Cote d'Ivoire"),'Country']='Ivory Coast'
CoTW_df.loc[(CoTW_df.Country=='Korea, North'),'Country']='North Korea'
CoTW_df.loc[(CoTW_df.Country=='Korea, South'),'Country']='South Korea'
CoTW_df.loc[(CoTW_df.Country=="Trinidad & Tobago"),'Country']='Trinidad and Tobago'
olymp2016medals.loc[(olymp2016medals.NOC=="Brazil*"),'NOC']='Brazil'
CoTW_country_names=CoTW_df['Country'].unique()
print(sorted(set(olymp2016medals['NOC'])-set(CoTW_country_names)) )
#print(sorted(set(CoTW_country_names)-set(olymp2016medals['NOC'])))
['Independent Olympic Athletes', 'Kosovo']
C:\Users\datta\anaconda3\lib\site-packages\pandas\core\indexing.py:1765: SettingWithCopyWarning: A value is trying to be set on a copy of a slice from a DataFrame. Try using .loc[row_indexer,col_indexer] = value instead See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy isetter(loc, value)
Need to `fix' all the anomalous data, particularly if we want to correlate the information across the data frames. Above, I did this manually.
Talking points:
- Challenges to do this at scale.
- Check for similarity of words: e.g., Korea, North vs North Korea; Trindad & Tobago vs Trinidad and Tobago
- What about Cote d'Ivoire vs Ivory Coast? Swaziland versus Switzerland? Niger vs Nigeria?
- What to do with United Kingdom vs Great Britain, Taiwan vs Chinese Taipei?
- Sometimes, you may be lucky and can leverage on Named entity recognition and matching tools.
Caveat: Different domains may be more or less amenable to certain solutions/heuristics.
Detour¶
For the particular problem at hand, namely, each country has been spelled variously, and we want to identify them to be equivalent, instead of writing our own tool, we may leverage on a nice tool someone already built!
Namely, country_converter: https://github.com/konstantinstadler/country_converter
There's also pycountry, which is useful in some related cases, but it maynot be suitable for our particular scenario (at least, I didn't figure out how to use it for our problem): https://pypi.org/project/pycountry/
#!pip install country_converter --upgrade
import country_converter as coco
# sanity check
some_names = ['Niger', 'Nigeria', 'Korea, North', "North Korea", "Cote d'Ivoire", "Ivory Coast", "Brazil*", "Brazil", "Great Britain", "United Kingdom", "Taiwan", "Chinese taipei", "Trinidad & Tobago", "Trinidad and Tobago", "Alice's Wonderland"]
standard_names = coco.convert(names=some_names, to='name_short')
print(standard_names)
Alice's Wonderland not found in regex
['Niger', 'Nigeria', 'North Korea', 'North Korea', "Cote d'Ivoire", "Cote d'Ivoire", 'Brazil', 'Brazil', 'United Kingdom', 'United Kingdom', 'Taiwan', 'Taiwan', 'Trinidad and Tobago', 'Trinidad and Tobago', 'not found']
Merge data frames¶
- how: {‘left’, ‘right’, ‘outer’, ‘inner’, ‘cross’}, default ‘inner’
- Pandas doc
- A nice discussion on various options how merger is done can be at Stack Overflow. [Below figure is from there.]
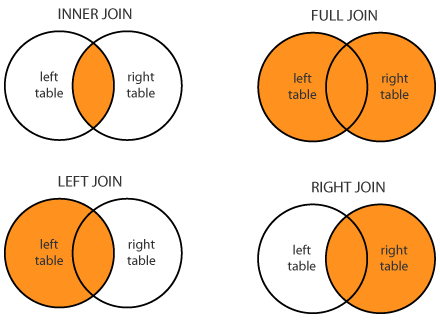
- Note that Pandas has also a Join method, which however in-fact join columns with other DataFrame either on index or on a key column. A good resource to check.
# Assuming that the data has been `cleaned'
# Determine all the countries with a population of more than 20 million
# as per the CountriesOfTheWorld table, that got no gold medals.
Merged_df=olymp2016medals.merge(CoTW_df[['Country','Population']],left_on='NOC', right_on='Country')
Merged_df[(Merged_df['Population']>20000000) & (Merged_df['Gold']==0)]
# Note: For the specific purpose, from CoTW_df, I do not need the other columns than Country and Population
# I could have cleaned the data more, e.g. NOC and Country columns are redundant.
| Rank | NOC | Gold | Silver | Bronze | Total | Country | Population | |
|---|---|---|---|---|---|---|---|---|
| 57 | 60 | Malaysia | 0 | 4 | 1 | 5 | Malaysia | 24385858 |
| 58 | 61 | Mexico | 0 | 3 | 2 | 5 | Mexico | 107449525 |
| 59 | 62 | Venezuela | 0 | 2 | 1 | 3 | Venezuela | 25730435 |
| 60 | 63 | Algeria | 0 | 2 | 0 | 2 | Algeria | 32930091 |
| 64 | 67 | India | 0 | 1 | 1 | 2 | India | 1095351995 |
| 69 | 69 | Philippines | 0 | 1 | 0 | 1 | Philippines | 89468677 |
| 72 | 75 | Egypt | 0 | 0 | 3 | 3 | Egypt | 78887007 |
| 79 | 78 | Morocco | 0 | 0 | 1 | 1 | Morocco | 33241259 |
| 80 | 78 | Nigeria | 0 | 0 | 1 | 1 | Nigeria | 131859731 |
Ungraded task 2.1: Ratio of total medals to per capita GDP
Determine the ratio of the number of total medals to the per capita GDP for each country as per the CountriesOfTheWorld table and sort them in decreasing order. Compare the results with others.
- Discuss: What subtleties are involved?
- How do you account for the countries which are not in the original Olympics table?
Groupby, Hierarchical index¶
# For each year for which data is available in the gap_df Dataframe
# What was the minimum recorded life expectancy?
gap_df.groupby(["year"])["lifeExp"].min()
year 1952 28.801 1957 30.332 1962 31.997 1967 34.020 1972 35.400 1977 31.220 1982 38.445 1987 39.906 1992 23.599 1997 36.087 2002 39.193 2007 39.613 Name: lifeExp, dtype: float64
# What if we also want to know, which country had said minimum life expectancy?
gap_df.loc[gap_df.groupby(["year"])["lifeExp"].idxmin()][['year','country','lifeExp']]
| year | country | lifeExp | |
|---|---|---|---|
| 0 | 1952 | Afghanistan | 28.801 |
| 1 | 1957 | Afghanistan | 30.332 |
| 2 | 1962 | Afghanistan | 31.997 |
| 3 | 1967 | Afghanistan | 34.020 |
| 1348 | 1972 | Sierra Leone | 35.400 |
| 221 | 1977 | Cambodia | 31.220 |
| 1350 | 1982 | Sierra Leone | 38.445 |
| 43 | 1987 | Angola | 39.906 |
| 1292 | 1992 | Rwanda | 23.599 |
| 1293 | 1997 | Rwanda | 36.087 |
| 1690 | 2002 | Zambia | 39.193 |
| 1463 | 2007 | Swaziland | 39.613 |
# Groupby can be useful in creating hierarchical indexing
# e.g., When we want to check the data by regions.
CoTW_df.groupby(['Region','Country']).mean()
# Many of the data, though representing numeric values, are in-fact treated as string in the current Dataframe.
# Refer to Ungraded Task 2.2 (below) for more on this. The nature of the data may limit what can be computed.
| Population | Area (sq. mi.) | GDP ($ per capita) | ||
|---|---|---|---|---|
| Region | Country | |||
| ASIA (EX. NEAR EAST) | Afghanistan | 31056997 | 647500 | 700.0 |
| Bangladesh | 147365352 | 144000 | 1900.0 | |
| Bhutan | 2279723 | 47000 | 1300.0 | |
| Brunei | 379444 | 5770 | 18600.0 | |
| Burma | 47382633 | 678500 | 1800.0 | |
| ... | ... | ... | ... | ... |
| WESTERN EUROPE | Portugal | 10605870 | 92391 | 18000.0 |
| San Marino | 29251 | 61 | 34600.0 | |
| Spain | 40397842 | 504782 | 22000.0 | |
| Sweden | 9016596 | 449964 | 26800.0 | |
| Switzerland | 7523934 | 41290 | 32700.0 |
227 rows × 3 columns
CoTW_df.groupby(['Region','Country']).max()
# With something like max, Pandas seems more generous in ignoring data type ;)
| Population | Area (sq. mi.) | Pop. Density (per sq. mi.) | Coastline (coast/area ratio) | Net migration | Infant mortality (per 1000 births) | GDP ($ per capita) | Literacy (%) | Phones (per 1000) | Arable (%) | Crops (%) | Other (%) | Climate | Birthrate | Deathrate | Agriculture | Industry | Service | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Country | ||||||||||||||||||
| ASIA (EX. NEAR EAST) | Afghanistan | 31056997 | 647500 | 48,0 | 0,00 | 23,06 | 163,07 | 700.0 | 36,0 | 3,2 | 12,13 | 0,22 | 87,65 | 1 | 46,6 | 20,34 | 0,38 | 0,24 | 0,38 |
| Bangladesh | 147365352 | 144000 | 1023,4 | 0,40 | -0,71 | 62,6 | 1900.0 | 43,1 | 7,3 | 62,11 | 3,07 | 34,82 | 2 | 29,8 | 8,27 | 0,199 | 0,198 | 0,603 | |
| Bhutan | 2279723 | 47000 | 48,5 | 0,00 | 0 | 100,44 | 1300.0 | 42,2 | 14,3 | 3,09 | 0,43 | 96,48 | 2 | 33,65 | 12,7 | 0,258 | 0,379 | 0,363 | |
| Brunei | 379444 | 5770 | 65,8 | 2,79 | 3,59 | 12,61 | 18600.0 | 93,9 | 237,2 | 0,57 | 0,76 | 98,67 | 2 | 18,79 | 3,45 | 0,036 | 0,561 | 0,403 | |
| Burma | 47382633 | 678500 | 69,8 | 0,28 | -1,8 | 67,24 | 1800.0 | 85,3 | 10,1 | 15,19 | 0,97 | 83,84 | 2 | 17,91 | 9,83 | 0,564 | 0,082 | 0,353 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| WESTERN EUROPE | Portugal | 10605870 | 92391 | 114,8 | 1,94 | 3,57 | 5,05 | 18000.0 | 93,3 | 399,2 | 21,75 | 7,81 | 70,44 | 3 | 10,72 | 10,5 | 0,053 | 0,274 | 0,673 |
| San Marino | 29251 | 61 | 479,5 | 0,00 | 10,98 | 5,73 | 34600.0 | 96,0 | 704,3 | 16,67 | 0 | 83,33 | NaN | 10,02 | 8,17 | NaN | NaN | NaN | |
| Spain | 40397842 | 504782 | 80,0 | 0,98 | 0,99 | 4,42 | 22000.0 | 97,9 | 453,5 | 26,07 | 9,87 | 64,06 | 3 | 10,06 | 9,72 | 0,04 | 0,295 | 0,665 | |
| Sweden | 9016596 | 449964 | 20,0 | 0,72 | 1,67 | 2,77 | 26800.0 | 99,0 | 715,0 | 6,54 | 0,01 | 93,45 | 3 | 10,27 | 10,31 | 0,011 | 0,282 | 0,707 | |
| Switzerland | 7523934 | 41290 | 182,2 | 0,00 | 4,05 | 4,39 | 32700.0 | 99,0 | 680,9 | 10,42 | 0,61 | 88,97 | 3 | 9,71 | 8,49 | 0,015 | 0,34 | 0,645 |
227 rows × 18 columns
# For what we want, an alternate, and simpler (and more robust, since we don't need to worry about data types)
# option is to just use create hierarchical index using set_index, and then sort_index.
CoTW_df.set_index(['Region','Country']).sort_index()
| Population | Area (sq. mi.) | Pop. Density (per sq. mi.) | Coastline (coast/area ratio) | Net migration | Infant mortality (per 1000 births) | GDP ($ per capita) | Literacy (%) | Phones (per 1000) | Arable (%) | Crops (%) | Other (%) | Climate | Birthrate | Deathrate | Agriculture | Industry | Service | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Country | ||||||||||||||||||
| ASIA (EX. NEAR EAST) | Afghanistan | 31056997 | 647500 | 48,0 | 0,00 | 23,06 | 163,07 | 700.0 | 36,0 | 3,2 | 12,13 | 0,22 | 87,65 | 1 | 46,6 | 20,34 | 0,38 | 0,24 | 0,38 |
| Bangladesh | 147365352 | 144000 | 1023,4 | 0,40 | -0,71 | 62,6 | 1900.0 | 43,1 | 7,3 | 62,11 | 3,07 | 34,82 | 2 | 29,8 | 8,27 | 0,199 | 0,198 | 0,603 | |
| Bhutan | 2279723 | 47000 | 48,5 | 0,00 | 0 | 100,44 | 1300.0 | 42,2 | 14,3 | 3,09 | 0,43 | 96,48 | 2 | 33,65 | 12,7 | 0,258 | 0,379 | 0,363 | |
| Brunei | 379444 | 5770 | 65,8 | 2,79 | 3,59 | 12,61 | 18600.0 | 93,9 | 237,2 | 0,57 | 0,76 | 98,67 | 2 | 18,79 | 3,45 | 0,036 | 0,561 | 0,403 | |
| Burma | 47382633 | 678500 | 69,8 | 0,28 | -1,8 | 67,24 | 1800.0 | 85,3 | 10,1 | 15,19 | 0,97 | 83,84 | 2 | 17,91 | 9,83 | 0,564 | 0,082 | 0,353 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| WESTERN EUROPE | Portugal | 10605870 | 92391 | 114,8 | 1,94 | 3,57 | 5,05 | 18000.0 | 93,3 | 399,2 | 21,75 | 7,81 | 70,44 | 3 | 10,72 | 10,5 | 0,053 | 0,274 | 0,673 |
| San Marino | 29251 | 61 | 479,5 | 0,00 | 10,98 | 5,73 | 34600.0 | 96,0 | 704,3 | 16,67 | 0 | 83,33 | NaN | 10,02 | 8,17 | NaN | NaN | NaN | |
| Spain | 40397842 | 504782 | 80,0 | 0,98 | 0,99 | 4,42 | 22000.0 | 97,9 | 453,5 | 26,07 | 9,87 | 64,06 | 3 | 10,06 | 9,72 | 0,04 | 0,295 | 0,665 | |
| Sweden | 9016596 | 449964 | 20,0 | 0,72 | 1,67 | 2,77 | 26800.0 | 99,0 | 715,0 | 6,54 | 0,01 | 93,45 | 3 | 10,27 | 10,31 | 0,011 | 0,282 | 0,707 | |
| Switzerland | 7523934 | 41290 | 182,2 | 0,00 | 4,05 | 4,39 | 32700.0 | 99,0 | 680,9 | 10,42 | 0,61 | 88,97 | 3 | 9,71 | 8,49 | 0,015 | 0,34 | 0,645 |
227 rows × 18 columns
Missing data¶
There are many reasons that data may be missing. e.g.:
- It may be inherently missing: not collected, not entered, etc.
- It may be implicitly missing.
- Missing entries may get created during data processing.
# load data (source: https://github.com/chendaniely/pandas_for_everyone)
pfe_path ='data/pandas_for_everyone_data/' # change this to adjust relative path
#visited_df = pd.read_csv(pfe_path+'survey_visited.csv')
#survey_df = pd.read_csv(pfe_path+'survey_survey.csv')
ebola_df= pd.read_csv(pfe_path+'country_timeseries.csv')
ebola_df.sample(6)
| Date | Day | Cases_Guinea | Cases_Liberia | Cases_SierraLeone | Cases_Nigeria | Cases_Senegal | Cases_UnitedStates | Cases_Spain | Cases_Mali | Deaths_Guinea | Deaths_Liberia | Deaths_SierraLeone | Deaths_Nigeria | Deaths_Senegal | Deaths_UnitedStates | Deaths_Spain | Deaths_Mali | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 108 | 4/14/2014 | 23 | 168.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 108.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 88 | 6/3/2014 | 73 | 344.0 | 13.0 | NaN | NaN | NaN | NaN | NaN | NaN | 215.0 | 12.0 | 6.0 | NaN | NaN | NaN | NaN | NaN |
| 100 | 4/24/2014 | 33 | NaN | 35.0 | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN |
| 35 | 10/19/2014 | 211 | 1540.0 | NaN | 3706.0 | 20.0 | 1.0 | 3.0 | 1.0 | NaN | 904.0 | NaN | 1259.0 | 8.0 | 0.0 | 1.0 | 0.0 | NaN |
| 22 | 11/11/2014 | 234 | 1919.0 | NaN | 5586.0 | 20.0 | 1.0 | 4.0 | 1.0 | 4.0 | 1166.0 | NaN | 1187.0 | 8.0 | 0.0 | 1.0 | 0.0 | 3.0 |
| 96 | 5/5/2014 | 44 | 235.0 | 13.0 | 0.0 | NaN | NaN | NaN | NaN | NaN | 157.0 | 11.0 | 0.0 | NaN | NaN | NaN | NaN | NaN |
# One option is to consider missing data to have some default value.
# e.g., in the case of number of Ebola cases, it may be fair to consider missing values to be 0.
ebola_df.fillna(0,inplace=True)
ebola_df
| Date | Day | Cases_Guinea | Cases_Liberia | Cases_SierraLeone | Cases_Nigeria | Cases_Senegal | Cases_UnitedStates | Cases_Spain | Cases_Mali | Deaths_Guinea | Deaths_Liberia | Deaths_SierraLeone | Deaths_Nigeria | Deaths_Senegal | Deaths_UnitedStates | Deaths_Spain | Deaths_Mali | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1/5/2015 | 289 | 2776.0 | 0.0 | 10030.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1786.0 | 0.0 | 2977.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1 | 1/4/2015 | 288 | 2775.0 | 0.0 | 9780.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1781.0 | 0.0 | 2943.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 | 1/3/2015 | 287 | 2769.0 | 8166.0 | 9722.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1767.0 | 3496.0 | 2915.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3 | 1/2/2015 | 286 | 0.0 | 8157.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3496.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 4 | 12/31/2014 | 284 | 2730.0 | 8115.0 | 9633.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1739.0 | 3471.0 | 2827.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 117 | 3/27/2014 | 5 | 103.0 | 8.0 | 6.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 66.0 | 6.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 118 | 3/26/2014 | 4 | 86.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 62.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 119 | 3/25/2014 | 3 | 86.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 60.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 120 | 3/24/2014 | 2 | 86.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 59.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 121 | 3/22/2014 | 0 | 49.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 29.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
122 rows × 18 columns
# Let's consider an example of how missing data may get created during the processing (because it was implicitly missing)
afg_df = gap_df[gap_df['country']=='Afghanistan'][['year','pop','gdpPercap']].tail(2).set_index('year')
afg_df
| pop | gdpPercap | |
|---|---|---|
| year | ||
| 2002 | 25268405 | 726.734055 |
| 2007 | 31889923 | 974.580338 |
# If we for some reason need yearly population and per capita GDP
# That information was missing implicitly, and during our processing, it may get so explicitly
afg_df=afg_df.reindex(afg_df.index.tolist() + list(range(2000, 2011)))
afg_df=afg_df[~afg_df.index.duplicated(keep='first')].sort_index()
afg_df
| pop | gdpPercap | |
|---|---|---|
| year | ||
| 2000 | NaN | NaN |
| 2001 | NaN | NaN |
| 2002 | 25268405.0 | 726.734055 |
| 2003 | NaN | NaN |
| 2004 | NaN | NaN |
| 2005 | NaN | NaN |
| 2006 | NaN | NaN |
| 2007 | 31889923.0 | 974.580338 |
| 2008 | NaN | NaN |
| 2009 | NaN | NaN |
| 2010 | NaN | NaN |
# forward fill (the population)
afg_df.loc[:,'pop'] = afg_df.loc[:,'pop'].ffill()
afg_df
| pop | gdpPercap | |
|---|---|---|
| year | ||
| 2000 | NaN | NaN |
| 2001 | NaN | NaN |
| 2002 | 25268405.0 | 726.734055 |
| 2003 | 25268405.0 | NaN |
| 2004 | 25268405.0 | NaN |
| 2005 | 25268405.0 | NaN |
| 2006 | 25268405.0 | NaN |
| 2007 | 31889923.0 | 974.580338 |
| 2008 | 31889923.0 | NaN |
| 2009 | 31889923.0 | NaN |
| 2010 | 31889923.0 | NaN |
# backward fill (the population)
afg_df.loc[:,'pop'] = afg_df.loc[:,'pop'].bfill()
afg_df
| pop | gdpPercap | |
|---|---|---|
| year | ||
| 2000 | 25268405.0 | NaN |
| 2001 | 25268405.0 | NaN |
| 2002 | 25268405.0 | 726.734055 |
| 2003 | 25268405.0 | NaN |
| 2004 | 25268405.0 | NaN |
| 2005 | 25268405.0 | NaN |
| 2006 | 25268405.0 | NaN |
| 2007 | 31889923.0 | 974.580338 |
| 2008 | 31889923.0 | NaN |
| 2009 | 31889923.0 | NaN |
| 2010 | 31889923.0 | NaN |
# interpolate fill: many caveats apply
# only linear interpolation is supported
# the gaps are filled with linear interpolation.
# the extremities just repeat the data
afg_df.loc[:,'gdpPercap'] = afg_df.loc[:,'gdpPercap'].interpolate(limit_direction='both')
afg_df
# you may need your own custom method
| pop | gdpPercap | |
|---|---|---|
| year | ||
| 2000 | 25268405.0 | 726.734055 |
| 2001 | 25268405.0 | 726.734055 |
| 2002 | 25268405.0 | 726.734055 |
| 2003 | 25268405.0 | 776.303312 |
| 2004 | 25268405.0 | 825.872568 |
| 2005 | 25268405.0 | 875.441825 |
| 2006 | 25268405.0 | 925.011082 |
| 2007 | 31889923.0 | 974.580338 |
| 2008 | 31889923.0 | 974.580338 |
| 2009 | 31889923.0 | 974.580338 |
| 2010 | 31889923.0 | 974.580338 |
# Be mindful regarding the semantics of the data
# This data is from a 'toy example' on video playbacks
# from Prof. Christopher Brooks' course on Data Science in Python
# Usage logs being collected from multiple users parallely, with some missing data
data_from_coursera_path ='data/coursera_sourced_data/'
log_df=pd.read_csv(data_from_coursera_path+'log.csv')
log_df
| time | user | video | playback position | paused | volume | |
|---|---|---|---|---|---|---|
| 0 | 1469974424 | cheryl | intro.html | 5 | False | 10.0 |
| 1 | 1469974454 | cheryl | intro.html | 6 | NaN | NaN |
| 2 | 1469974544 | cheryl | intro.html | 9 | NaN | NaN |
| 3 | 1469974574 | cheryl | intro.html | 10 | NaN | NaN |
| 4 | 1469977514 | bob | intro.html | 1 | NaN | NaN |
| 5 | 1469977544 | bob | intro.html | 1 | NaN | NaN |
| 6 | 1469977574 | bob | intro.html | 1 | NaN | NaN |
| 7 | 1469977604 | bob | intro.html | 1 | NaN | NaN |
| 8 | 1469974604 | cheryl | intro.html | 11 | NaN | NaN |
| 9 | 1469974694 | cheryl | intro.html | 14 | NaN | NaN |
| 10 | 1469974724 | cheryl | intro.html | 15 | NaN | NaN |
| 11 | 1469974454 | sue | advanced.html | 24 | NaN | NaN |
| 12 | 1469974524 | sue | advanced.html | 25 | NaN | NaN |
| 13 | 1469974424 | sue | advanced.html | 23 | False | 10.0 |
| 14 | 1469974554 | sue | advanced.html | 26 | NaN | NaN |
| 15 | 1469974624 | sue | advanced.html | 27 | NaN | NaN |
| 16 | 1469974654 | sue | advanced.html | 28 | NaN | 5.0 |
| 17 | 1469974724 | sue | advanced.html | 29 | NaN | NaN |
| 18 | 1469974484 | cheryl | intro.html | 7 | NaN | NaN |
| 19 | 1469974514 | cheryl | intro.html | 8 | NaN | NaN |
| 20 | 1469974754 | sue | advanced.html | 30 | NaN | NaN |
| 21 | 1469974824 | sue | advanced.html | 31 | NaN | NaN |
| 22 | 1469974854 | sue | advanced.html | 32 | NaN | NaN |
| 23 | 1469974924 | sue | advanced.html | 33 | NaN | NaN |
| 24 | 1469977424 | bob | intro.html | 1 | True | 10.0 |
| 25 | 1469977454 | bob | intro.html | 1 | NaN | NaN |
| 26 | 1469977484 | bob | intro.html | 1 | NaN | NaN |
| 27 | 1469977634 | bob | intro.html | 1 | NaN | NaN |
| 28 | 1469977664 | bob | intro.html | 1 | NaN | NaN |
| 29 | 1469974634 | cheryl | intro.html | 12 | NaN | NaN |
| 30 | 1469974664 | cheryl | intro.html | 13 | NaN | NaN |
| 31 | 1469977694 | bob | intro.html | 1 | NaN | NaN |
| 32 | 1469977724 | bob | intro.html | 1 | NaN | NaN |
# Let's view the data sorted by time
log_df=log_df.set_index('time').sort_index()
log_df
| user | video | playback position | paused | volume | |
|---|---|---|---|---|---|
| time | |||||
| 1469974424 | cheryl | intro.html | 5 | False | 10.0 |
| 1469974424 | sue | advanced.html | 23 | False | 10.0 |
| 1469974454 | cheryl | intro.html | 6 | NaN | NaN |
| 1469974454 | sue | advanced.html | 24 | NaN | NaN |
| 1469974484 | cheryl | intro.html | 7 | NaN | NaN |
| 1469974514 | cheryl | intro.html | 8 | NaN | NaN |
| 1469974524 | sue | advanced.html | 25 | NaN | NaN |
| 1469974544 | cheryl | intro.html | 9 | NaN | NaN |
| 1469974554 | sue | advanced.html | 26 | NaN | NaN |
| 1469974574 | cheryl | intro.html | 10 | NaN | NaN |
| 1469974604 | cheryl | intro.html | 11 | NaN | NaN |
| 1469974624 | sue | advanced.html | 27 | NaN | NaN |
| 1469974634 | cheryl | intro.html | 12 | NaN | NaN |
| 1469974654 | sue | advanced.html | 28 | NaN | 5.0 |
| 1469974664 | cheryl | intro.html | 13 | NaN | NaN |
| 1469974694 | cheryl | intro.html | 14 | NaN | NaN |
| 1469974724 | cheryl | intro.html | 15 | NaN | NaN |
| 1469974724 | sue | advanced.html | 29 | NaN | NaN |
| 1469974754 | sue | advanced.html | 30 | NaN | NaN |
| 1469974824 | sue | advanced.html | 31 | NaN | NaN |
| 1469974854 | sue | advanced.html | 32 | NaN | NaN |
| 1469974924 | sue | advanced.html | 33 | NaN | NaN |
| 1469977424 | bob | intro.html | 1 | True | 10.0 |
| 1469977454 | bob | intro.html | 1 | NaN | NaN |
| 1469977484 | bob | intro.html | 1 | NaN | NaN |
| 1469977514 | bob | intro.html | 1 | NaN | NaN |
| 1469977544 | bob | intro.html | 1 | NaN | NaN |
| 1469977574 | bob | intro.html | 1 | NaN | NaN |
| 1469977604 | bob | intro.html | 1 | NaN | NaN |
| 1469977634 | bob | intro.html | 1 | NaN | NaN |
| 1469977664 | bob | intro.html | 1 | NaN | NaN |
| 1469977694 | bob | intro.html | 1 | NaN | NaN |
| 1469977724 | bob | intro.html | 1 | NaN | NaN |
log_df=log_df.reset_index().set_index(['user','time']).sort_index()
# We want to use a 2-level index, so that all entries for a given user are together
# Chaining can keep your code short. Make sure it does not make it too cryptic though ;)
# Motivation: Using user Cheryl's volume setting to fill-in user Bob's volume may not be meaningful
log_df
| video | playback position | paused | volume | ||
|---|---|---|---|---|---|
| user | time | ||||
| bob | 1469977424 | intro.html | 1 | True | 10.0 |
| 1469977454 | intro.html | 1 | NaN | NaN | |
| 1469977484 | intro.html | 1 | NaN | NaN | |
| 1469977514 | intro.html | 1 | NaN | NaN | |
| 1469977544 | intro.html | 1 | NaN | NaN | |
| 1469977574 | intro.html | 1 | NaN | NaN | |
| 1469977604 | intro.html | 1 | NaN | NaN | |
| 1469977634 | intro.html | 1 | NaN | NaN | |
| 1469977664 | intro.html | 1 | NaN | NaN | |
| 1469977694 | intro.html | 1 | NaN | NaN | |
| 1469977724 | intro.html | 1 | NaN | NaN | |
| cheryl | 1469974424 | intro.html | 5 | False | 10.0 |
| 1469974454 | intro.html | 6 | NaN | NaN | |
| 1469974484 | intro.html | 7 | NaN | NaN | |
| 1469974514 | intro.html | 8 | NaN | NaN | |
| 1469974544 | intro.html | 9 | NaN | NaN | |
| 1469974574 | intro.html | 10 | NaN | NaN | |
| 1469974604 | intro.html | 11 | NaN | NaN | |
| 1469974634 | intro.html | 12 | NaN | NaN | |
| 1469974664 | intro.html | 13 | NaN | NaN | |
| 1469974694 | intro.html | 14 | NaN | NaN | |
| 1469974724 | intro.html | 15 | NaN | NaN | |
| sue | 1469974424 | advanced.html | 23 | False | 10.0 |
| 1469974454 | advanced.html | 24 | NaN | NaN | |
| 1469974524 | advanced.html | 25 | NaN | NaN | |
| 1469974554 | advanced.html | 26 | NaN | NaN | |
| 1469974624 | advanced.html | 27 | NaN | NaN | |
| 1469974654 | advanced.html | 28 | NaN | 5.0 | |
| 1469974724 | advanced.html | 29 | NaN | NaN | |
| 1469974754 | advanced.html | 30 | NaN | NaN | |
| 1469974824 | advanced.html | 31 | NaN | NaN | |
| 1469974854 | advanced.html | 32 | NaN | NaN | |
| 1469974924 | advanced.html | 33 | NaN | NaN |
# Now, it makes certain sense to assume that
# the missing values correspond to the last recorded value for that particular user
# Forward the Fill! (based on a reasonable Foundation of the assumptions ;)
log_df = log_df.fillna(method='ffill')
log_df
| video | playback position | paused | volume | ||
|---|---|---|---|---|---|
| user | time | ||||
| bob | 1469977424 | intro.html | 1 | True | 10.0 |
| 1469977454 | intro.html | 1 | True | 10.0 | |
| 1469977484 | intro.html | 1 | True | 10.0 | |
| 1469977514 | intro.html | 1 | True | 10.0 | |
| 1469977544 | intro.html | 1 | True | 10.0 | |
| 1469977574 | intro.html | 1 | True | 10.0 | |
| 1469977604 | intro.html | 1 | True | 10.0 | |
| 1469977634 | intro.html | 1 | True | 10.0 | |
| 1469977664 | intro.html | 1 | True | 10.0 | |
| 1469977694 | intro.html | 1 | True | 10.0 | |
| 1469977724 | intro.html | 1 | True | 10.0 | |
| cheryl | 1469974424 | intro.html | 5 | False | 10.0 |
| 1469974454 | intro.html | 6 | False | 10.0 | |
| 1469974484 | intro.html | 7 | False | 10.0 | |
| 1469974514 | intro.html | 8 | False | 10.0 | |
| 1469974544 | intro.html | 9 | False | 10.0 | |
| 1469974574 | intro.html | 10 | False | 10.0 | |
| 1469974604 | intro.html | 11 | False | 10.0 | |
| 1469974634 | intro.html | 12 | False | 10.0 | |
| 1469974664 | intro.html | 13 | False | 10.0 | |
| 1469974694 | intro.html | 14 | False | 10.0 | |
| 1469974724 | intro.html | 15 | False | 10.0 | |
| sue | 1469974424 | advanced.html | 23 | False | 10.0 |
| 1469974454 | advanced.html | 24 | False | 10.0 | |
| 1469974524 | advanced.html | 25 | False | 10.0 | |
| 1469974554 | advanced.html | 26 | False | 10.0 | |
| 1469974624 | advanced.html | 27 | False | 10.0 | |
| 1469974654 | advanced.html | 28 | False | 5.0 | |
| 1469974724 | advanced.html | 29 | False | 5.0 | |
| 1469974754 | advanced.html | 30 | False | 5.0 | |
| 1469974824 | advanced.html | 31 | False | 5.0 | |
| 1469974854 | advanced.html | 32 | False | 5.0 | |
| 1469974924 | advanced.html | 33 | False | 5.0 |
Some basic text processing & RegEx¶
# Tokens
NTU_tweet ="""Doctors may soon identify #diabetes patients who have a higher risk of vascular #inflammation with a one-step process using a ‘lab-on-a-chip’ device developed by scientists from @NTUsg, @TTSH and @MIT. #NTUsg2025 #NTUsgResearch https://bit.ly/ExoDFFchip"""
Tweet_tokens=NTU_tweet.split()
print(Tweet_tokens)
['Doctors', 'may', 'soon', 'identify', '#diabetes', 'patients', 'who', 'have', 'a', 'higher', 'risk', 'of', 'vascular', '#inflammation', 'with', 'a', 'one-step', 'process', 'using', 'a', '‘lab-on-a-chip’', 'device', 'developed', 'by', 'scientists', 'from', '@NTUsg,', '@TTSH', 'and', '@MIT.', '#NTUsg2025', '#NTUsgResearch', 'https://bit.ly/ExoDFFchip']
# Find 'long' words (more than 4 letters) which are not hashtags, call outs, or urls!
Tweet_longwords=[x for x in Tweet_tokens if (len(x)>4) & (x.find('#')<0) & (x.find('@')<0) & (x.find('://')<0)]
print(Tweet_longwords)
['Doctors', 'identify', 'patients', 'higher', 'vascular', 'one-step', 'process', 'using', '‘lab-on-a-chip’', 'device', 'developed', 'scientists']
# What about, if we want to find all the hashtags?
# Attempt 1:
Tweet_hashtags1a=[x for x in Tweet_tokens if (x.find('#')>=0) ]
Tweet_tokens.append("stranger#things")
Tweet_hashtags1b=[x for x in Tweet_tokens if (x.find('#')>=0) ]
print(Tweet_hashtags1a)
print(Tweet_hashtags1b)
['#diabetes', '#inflammation', '#NTUsg2025', '#NTUsgResearch'] ['#diabetes', '#inflammation', '#NTUsg2025', '#NTUsgResearch', 'stranger#things']
# Attempt 2:
# Just in case there's something like `stranger#things' which is not a hashtag!
# We might want to use one of the following:
Tweet_hashtags2=[x for x in Tweet_tokens if (x.find('#')==0) ]
Tweet_hashtags3=[x for x in Tweet_tokens if x.startswith('#') ]
print(Tweet_hashtags2)
print(Tweet_hashtags3)
['#diabetes', '#inflammation', '#NTUsg2025', '#NTUsgResearch'] ['#diabetes', '#inflammation', '#NTUsg2025', '#NTUsgResearch']
# Find all mentions of 'NTU'
Contains_NTU1=[x for x in Tweet_tokens if (x.find('NTU')>=0) ]
Contains_NTU2=[x for x in Tweet_tokens if 'NTU' in x]
print(Contains_NTU1)
print(Contains_NTU2)
['@NTUsg,', '#NTUsg2025', '#NTUsgResearch'] ['@NTUsg,', '#NTUsg2025', '#NTUsgResearch']
Some frequently used & useful string methods
- checking: isalpha/isdigit/isalnum
- transforming: lower/upper/title/replace/strip/rstrip
- locate: find/rfind
See [https://docs.python.org/3/library/stdtypes.html] for a more exhaustive list.
# Let's redo the extraction of hashtags with a RegEx
[x for x in Tweet_tokens if re.search('\B#[A-Za-z-0-9_]+',x)]
# For the specifc example tweet, if we used re.search('#',x), it would have sufficed.
# But then it would not have managed to handle the 'stranger' cases such as isolated #, ## or hash sandwiched by text..
['#diabetes', '#inflammation', '#NTUsg2025', '#NTUsgResearch']
How to interpret '\B#[A-Za-z-0-9_]+'?¶
[x for x in Tweet_tokens if re.search('\B#[A-Za-z-0-9_]+',x)]
\B Matches boundary with non-alphanumeric characters
[A-Za-z-0-9_] Matches any of A to Z, a to z, 0 to 9 and _
+ Greedily matches that expression on its immediate left is present at least once.
# The same RegEx can be rewritten in a much shorter (and slightly cryptic) manner
[x for x in Tweet_tokens if re.search('\B#\w+',x)]
['#diabetes', '#inflammation', '#NTUsg2025', '#NTUsgResearch']
# Open a file, read it line-by-line and create a Data Series
# The following data is obtained from Prof V. Vydiswaran's course on Applied Text Mining in Python
doc = []
with open(data_from_coursera_path+'text-with-dates.txt','r') as file:
for line in file:
doc.append(line.rstrip()) # used rstrip to remove trailing newlines \n
datetxt_df = pd.Series(doc)
datetxt_df
0 03/25/93 Total time of visit (in minutes):
1 6/18/85 Primary Care Doctor:
2 sshe plans to move as of 7/8/71 In-Home Servic...
3 7 on 9/27/75 Audit C Score Current:
4 2/6/96 sleep studyPain Treatment Pain Level (N...
...
495 1979 Family Psych History: Family History of S...
496 therapist and friend died in ~2006 Parental/Ca...
497 2008 partial thyroidectomy
498 sPt describes a history of sexual abuse as a c...
499 . In 1980, patient was living in Naples and de...
Length: 500, dtype: object
# What does the following code do?
[x for x in datetxt_df if re.findall(r'\d{1,2}[/|-]\d{1,2}[/|-]\d{2,4}',x)]
['03/25/93 Total time of visit (in minutes):', '6/18/85 Primary Care Doctor:', 'sshe plans to move as of 7/8/71 In-Home Services: None', '7 on 9/27/75 Audit C Score Current:', '2/6/96 sleep studyPain Treatment Pain Level (Numeric Scale): 7', '.Per 7/06/79 Movement D/O note:', "4, 5/18/78 Patient's thoughts about current substance abuse:", '10/24/89 CPT Code: 90801 - Psychiatric Diagnosis Interview', '3/7/86 SOS-10 Total Score:', '(4/10/71)Score-1Audit C Score Current:', '(5/11/85) Crt-1.96, BUN-26; AST/ALT-16/22; WBC_12.6Activities of Daily Living (ADL) Bathing: Independent', '4/09/75 SOS-10 Total Score:', '8/01/98 Communication with referring physician?: Done', '1/26/72 Communication with referring physician?: Not Done', '5/24/1990 CPT Code: 90792: With medical services', '1/25/2011 CPT Code: 90792: With medical services', '4/12/82 Total time of visit (in minutes):', '1; 10/13/1976 Audit C Score, Highest/Date:', '4, 4/24/98 Relevant Drug History:', ') 59 yo unemployed w referred by Urgent Care for psychiatric evaluation and follow up. The patient reports she has been dx w BAD. Her main complaint today is anergy. Ms. Hartman was evaluated on one occasion 5/21/77. She was a cooperative but somewhat vague historian.History of Present Illness and Precipitating Events', '7/21/98 Total time of visit (in minutes):', '10/21/79 SOS-10 Total Score:', '3/03/90 CPT Code: 90792: With medical services', '2/11/76 CPT Code: 90792: With medical services', '07/25/1984 CPT Code: 90791: No medical services', '4-13-82 Other Child Mental Health Outcomes Scales Used:', '9/22/89 CPT Code: 90792: With medical services', '9/02/76 CPT Code: 90791: No medical services', '9/12/71 [report_end]', '10/24/86 Communication with referring physician?: Done', '03/31/1985 Total time of visit (in minutes):', '7/20/72 CPT Code: 90791: No medical services', '4/12/87= 1, negativeAudit C Score Current:', '06/20/91 Total time of visit (in minutes):', '5/12/2012 Primary Care Doctor:', '3/15/83 SOS-10 Total Score:', '2/14/73 CPT Code: 90801 - Psychiatric Diagnosis Interview', '5/24/88 CPT Code: 90792: With medical services', '7/27/1986 Total time of visit (in minutes):', '1-14-81 Communication with referring physician?: Done', '7-29-75 CPT Code: 90801 - Psychiatric Diagnosis Interview', '(6/24/87) TSH-2.18; Activities of Daily Living (ADL) Bathing: Independent', '8/14/94 Primary Care Doctor:', '4/13/2002 Primary Care Doctor:', '8/16/82 CPT Code: 90792: With medical services', '2/15/1998 Total time of visit (in minutes):', '7/15/91 CPT Code: 90792: With medical services', '06/12/94 SOS-10 Total Score:', '9/17/84 Communication with referring physician?: Done', '2/28/75 Other Adult Mental Health Outcomes Scales Used:', 'sOP WPM - Dr. Romo-psychopharm since 11/22/75', '"In the context of the patient\'s work up, she had Neuropsychological testing 5/24/91. She was referred because of her cognitive decline and symptoms of motor dysfunction to help characterize her current functioning. :', '6/13/92 CPT Code: 90791: No medical services', '7/11/71 SOS-10 Total Score:', '12/26/86 CPT Code: 90791: No medical services', '10/11/1987 CPT Code: 90791: No medical services', '3/14/95 Primary Care Doctor:', '12/01/73 CPT Code: 90791: No medical services', '0: 12/5/2010 Audit C Score Current:', '08/20/1982 SOS-10 Total Score:', '7/24/95 SOS-10 Total Score:', '8/06/83 CPT Code: 90791: No medical services', '02/22/92 SOS-10 Total Score:', '6/28/87 Impression Strengths/Abilities:', '07/29/1994 CPT code: 99203', '08/11/78 CPT Code: 90801 - Psychiatric Diagnosis Interview', '10/29/91 Communication with referring physician?: Done', '7/6/91 SOS-10 Total Score:', 'onone as of 1/21/87 Protective Factors:', '24 yo right handed woman with history of large right frontal mass s/p resection 11/3/1985 who had recent urgent R cranial wound revision and placement of L EVD for declining vision and increased drainage from craniotomy incision site and possible infection. She has a hx of secondary mania related to psychosis and manipulation of her right frontal lobe.', '7/04/82 CPT Code: 90801 - Psychiatric Diagnosis Interview', '4-13-89 Communication with referring physician?: Not Done', 'Lithium 0.25 (7/11/77). LFTS wnl. Urine tox neg. Serum tox + fluoxetine 500; otherwise neg. TSH 3.28. BUN/Cr: 16/0.83. Lipids unremarkable. B12 363, Folate >20. CBC: 4.9/36/308 Pertinent Medical Review of Systems Constitutional:', '4/12/74= 1Caffeine / Tobacco Use Caffeinated products: Yes', '09/19/81 CPT Code: 90792: With medical services', '9/6/79 Primary Care Doctor:', '12/5/87 Total time of visit (in minutes):', '01/05/1999 [report_end]', '4/22/80 SOS-10 Total Score:', '10/04/98 SOS-10 Total Score:', '.Mr. Echeverria described having panic-like experiences on at least three occasions. The first of these episodes occurred when facial meshing was placed on him during radiation therapy. He recalled the experience vividly and described "freaking out", involving a sensation of crushing in his chest, difficulty breathing, a fear that he could die, and intense emotional disquiet. The second episode occurred during the last 10-15 minutes of an MRI scan. Mr. Echeverria noted feeling surprised and fearful when the feelings of panic started. Similar physiological and psychological symptoms were reported as above. The third episode occurred recently on 6/29/81. Mr.Echeverria was driving his car and felt similar symptoms as described above. He reported fearing that he would crash his vehicle and subsequently pulled over and abandoned his vehicle.', '. Wile taking additional history, pt endorsed moderate depression and anxiety last 2 weeks, even though her period was on 8/04/78. Pt then acknowledged low to moderate mood and anxiety symptoms throughout the month, with premenstrual worsening. This mood pattern seems to have existed for many years, while premenstrual worsening increased in severity during last ~ 2 years, in context of likely peri menopause transition. Pt reported average of 3 hot flashes per day and 2 night sweats.', 'Death of mother; 7/07/1974 Meaningful activities/supports:', '09/14/2000 CPT Code: 90792: With medical services', '5/18/71 Total time of visit (in minutes):', '8/09/1981 Communication with referring physician?: Done', '6/05/93 CPT Code: 90791: No medical services', ')Dilantin (PHENYTOIN) 100 MG CAPSULE Take 2 Capsule(s) PO HS; No Change (Taking Differently), Comments: decreased from 290 daily to 260 mg daily due to elevated phenytoin level from 8/9/97', '12/8/82 Audit C=3Audit C Score Current:', '8/26/89 CPT Code: 90791: No medical services', '10/13/95 CPT Code: 90791: No medical services', '4/19/91 Communication with referring physician?: Not Done', '.APS - Psychiatry consult paged/requested in person at 04/08/2004 16:39 Patrick, Christian [hpp2]', '9/20/76 CPT Code: 90801 - Psychiatric Diagnosis Interview', '12/08/1990 @11 am [report_end]', '4/11/1974 Chief Complaint / HPI Chief Complaint (Patients own words)', '7/18/86 SOS-10 Total Score:', '3/31/91 Communication with referring physician?: Done', '5/13/72 Other Adult Mental Health Outcomes Scales Used:', '011/14/83 Audit C Score Current:', '8/16/92 SOS-10 Total Score:', '10/05/97 CPT Code: 90791: No medical services', '07/18/2002 CPT Code: 90792: With medical services', '9/22/82 Total time of visit (in minutes):', '2/24/74 SOS-10 Total Score:', '(2/03/78) TSH-0.90 Activities of Daily Living (ADL) Bathing: Independent', '2/11/2006 CPT Code: 90791: No medical services', "Pt is a 21 year old, single, heterosexual identified, Liechtenstein male. He resides in Rabat, in a rented apartment, with a roommate. He has not graduated from high school nor has he earned his GED. He has been working full time, steadily, since the age of 16. He recently left a job at jacobs engineering group where he worked for about 5 years. He started working at becton dickinson a few weeks ago. Referral to SMH was precipitated by pt seeing his PCP, Dr. Ford, for a physical and sharing concerns re: depression. He was having suicidal thoughts the week of 8/22/83 and noticed that his depression and anxiety symptoms were starting to interfere with work. Per Dr. Ford's recommendation, he contacted PAL for assistance. When screened for substance use, pt was referred to SMH for an intake. Pt was scheduled to be seen last week due to urgency of his depression and SI, but missed the intake appointment due to getting extremely intoxicated the night before. Pt presents today seeking individual therapy and psychiatry services to address longstanding depression.", 'PET Scan (DPSH 5/04/74): 1) Marked hypometabolism involving the bilateral caudate nuclei. This is a non-specific finding, although this raises the question of a multisystem atrophy, specifically MSA-P (striatonigral degeneration), 2) Cortical hypometabolism involving the right inferior frontal gyrus. This is a non-specific finding of uncertain significance, and is not definitively related to a neurodegenerative process, although pathology in this region is not excluded', '7/20/2011 [report_end]', '6/17/95 Total time of visit (in minutes):', '6/10/72 SOS-10 Total Score:', 'nPt denied use to me but endorsed use on 10/16/82 as part of BVH initial visit summary supplement by Nicholas BenjaminOpiates: Yes', '12/15/92 CPT Code: 90801 - Psychiatric Diagnosis Interview', '12/8/97 SOS-10 Total Score:', '4/05/89 Primary Care Doctor:', '12/04/87 SOS-10 Total Score:', '4 (6/20/77)Audit C Score Current:', 'see 4/27/2006 consult note Dr. GuevaraWhat factors in prior treatment were helpful/not helpful:', '07/17/92 CPT Code: 90791: No medical services', '12/22/98 CPT Code: 90801 - Psychiatric Diagnosis Interview', '10/02/96 Age:', '11/05/90 CPT Code: 90792: With medical services', '5/04/77 CPT Code: 90792: With medical services', '2/27/96 Communication with referring physician?: Done']
[x for x in datetxt_df if re.search(r'\d{1,2}[/|-]\d{1,2}[/|-]\d{2,4}',x)]
# Search returns the first occurrence, while findall finds all the (non-overlapping) matches.
# For our usage here, either suffices.
# Even re.match() would suffice for the current purpose.
# Check the subtle differences between match/search/findall for yourself!
['03/25/93 Total time of visit (in minutes):', '6/18/85 Primary Care Doctor:', 'sshe plans to move as of 7/8/71 In-Home Services: None', '7 on 9/27/75 Audit C Score Current:', '2/6/96 sleep studyPain Treatment Pain Level (Numeric Scale): 7', '.Per 7/06/79 Movement D/O note:', "4, 5/18/78 Patient's thoughts about current substance abuse:", '10/24/89 CPT Code: 90801 - Psychiatric Diagnosis Interview', '3/7/86 SOS-10 Total Score:', '(4/10/71)Score-1Audit C Score Current:', '(5/11/85) Crt-1.96, BUN-26; AST/ALT-16/22; WBC_12.6Activities of Daily Living (ADL) Bathing: Independent', '4/09/75 SOS-10 Total Score:', '8/01/98 Communication with referring physician?: Done', '1/26/72 Communication with referring physician?: Not Done', '5/24/1990 CPT Code: 90792: With medical services', '1/25/2011 CPT Code: 90792: With medical services', '4/12/82 Total time of visit (in minutes):', '1; 10/13/1976 Audit C Score, Highest/Date:', '4, 4/24/98 Relevant Drug History:', ') 59 yo unemployed w referred by Urgent Care for psychiatric evaluation and follow up. The patient reports she has been dx w BAD. Her main complaint today is anergy. Ms. Hartman was evaluated on one occasion 5/21/77. She was a cooperative but somewhat vague historian.History of Present Illness and Precipitating Events', '7/21/98 Total time of visit (in minutes):', '10/21/79 SOS-10 Total Score:', '3/03/90 CPT Code: 90792: With medical services', '2/11/76 CPT Code: 90792: With medical services', '07/25/1984 CPT Code: 90791: No medical services', '4-13-82 Other Child Mental Health Outcomes Scales Used:', '9/22/89 CPT Code: 90792: With medical services', '9/02/76 CPT Code: 90791: No medical services', '9/12/71 [report_end]', '10/24/86 Communication with referring physician?: Done', '03/31/1985 Total time of visit (in minutes):', '7/20/72 CPT Code: 90791: No medical services', '4/12/87= 1, negativeAudit C Score Current:', '06/20/91 Total time of visit (in minutes):', '5/12/2012 Primary Care Doctor:', '3/15/83 SOS-10 Total Score:', '2/14/73 CPT Code: 90801 - Psychiatric Diagnosis Interview', '5/24/88 CPT Code: 90792: With medical services', '7/27/1986 Total time of visit (in minutes):', '1-14-81 Communication with referring physician?: Done', '7-29-75 CPT Code: 90801 - Psychiatric Diagnosis Interview', '(6/24/87) TSH-2.18; Activities of Daily Living (ADL) Bathing: Independent', '8/14/94 Primary Care Doctor:', '4/13/2002 Primary Care Doctor:', '8/16/82 CPT Code: 90792: With medical services', '2/15/1998 Total time of visit (in minutes):', '7/15/91 CPT Code: 90792: With medical services', '06/12/94 SOS-10 Total Score:', '9/17/84 Communication with referring physician?: Done', '2/28/75 Other Adult Mental Health Outcomes Scales Used:', 'sOP WPM - Dr. Romo-psychopharm since 11/22/75', '"In the context of the patient\'s work up, she had Neuropsychological testing 5/24/91. She was referred because of her cognitive decline and symptoms of motor dysfunction to help characterize her current functioning. :', '6/13/92 CPT Code: 90791: No medical services', '7/11/71 SOS-10 Total Score:', '12/26/86 CPT Code: 90791: No medical services', '10/11/1987 CPT Code: 90791: No medical services', '3/14/95 Primary Care Doctor:', '12/01/73 CPT Code: 90791: No medical services', '0: 12/5/2010 Audit C Score Current:', '08/20/1982 SOS-10 Total Score:', '7/24/95 SOS-10 Total Score:', '8/06/83 CPT Code: 90791: No medical services', '02/22/92 SOS-10 Total Score:', '6/28/87 Impression Strengths/Abilities:', '07/29/1994 CPT code: 99203', '08/11/78 CPT Code: 90801 - Psychiatric Diagnosis Interview', '10/29/91 Communication with referring physician?: Done', '7/6/91 SOS-10 Total Score:', 'onone as of 1/21/87 Protective Factors:', '24 yo right handed woman with history of large right frontal mass s/p resection 11/3/1985 who had recent urgent R cranial wound revision and placement of L EVD for declining vision and increased drainage from craniotomy incision site and possible infection. She has a hx of secondary mania related to psychosis and manipulation of her right frontal lobe.', '7/04/82 CPT Code: 90801 - Psychiatric Diagnosis Interview', '4-13-89 Communication with referring physician?: Not Done', 'Lithium 0.25 (7/11/77). LFTS wnl. Urine tox neg. Serum tox + fluoxetine 500; otherwise neg. TSH 3.28. BUN/Cr: 16/0.83. Lipids unremarkable. B12 363, Folate >20. CBC: 4.9/36/308 Pertinent Medical Review of Systems Constitutional:', '4/12/74= 1Caffeine / Tobacco Use Caffeinated products: Yes', '09/19/81 CPT Code: 90792: With medical services', '9/6/79 Primary Care Doctor:', '12/5/87 Total time of visit (in minutes):', '01/05/1999 [report_end]', '4/22/80 SOS-10 Total Score:', '10/04/98 SOS-10 Total Score:', '.Mr. Echeverria described having panic-like experiences on at least three occasions. The first of these episodes occurred when facial meshing was placed on him during radiation therapy. He recalled the experience vividly and described "freaking out", involving a sensation of crushing in his chest, difficulty breathing, a fear that he could die, and intense emotional disquiet. The second episode occurred during the last 10-15 minutes of an MRI scan. Mr. Echeverria noted feeling surprised and fearful when the feelings of panic started. Similar physiological and psychological symptoms were reported as above. The third episode occurred recently on 6/29/81. Mr.Echeverria was driving his car and felt similar symptoms as described above. He reported fearing that he would crash his vehicle and subsequently pulled over and abandoned his vehicle.', '. Wile taking additional history, pt endorsed moderate depression and anxiety last 2 weeks, even though her period was on 8/04/78. Pt then acknowledged low to moderate mood and anxiety symptoms throughout the month, with premenstrual worsening. This mood pattern seems to have existed for many years, while premenstrual worsening increased in severity during last ~ 2 years, in context of likely peri menopause transition. Pt reported average of 3 hot flashes per day and 2 night sweats.', 'Death of mother; 7/07/1974 Meaningful activities/supports:', '09/14/2000 CPT Code: 90792: With medical services', '5/18/71 Total time of visit (in minutes):', '8/09/1981 Communication with referring physician?: Done', '6/05/93 CPT Code: 90791: No medical services', ')Dilantin (PHENYTOIN) 100 MG CAPSULE Take 2 Capsule(s) PO HS; No Change (Taking Differently), Comments: decreased from 290 daily to 260 mg daily due to elevated phenytoin level from 8/9/97', '12/8/82 Audit C=3Audit C Score Current:', '8/26/89 CPT Code: 90791: No medical services', '10/13/95 CPT Code: 90791: No medical services', '4/19/91 Communication with referring physician?: Not Done', '.APS - Psychiatry consult paged/requested in person at 04/08/2004 16:39 Patrick, Christian [hpp2]', '9/20/76 CPT Code: 90801 - Psychiatric Diagnosis Interview', '12/08/1990 @11 am [report_end]', '4/11/1974 Chief Complaint / HPI Chief Complaint (Patients own words)', '7/18/86 SOS-10 Total Score:', '3/31/91 Communication with referring physician?: Done', '5/13/72 Other Adult Mental Health Outcomes Scales Used:', '011/14/83 Audit C Score Current:', '8/16/92 SOS-10 Total Score:', '10/05/97 CPT Code: 90791: No medical services', '07/18/2002 CPT Code: 90792: With medical services', '9/22/82 Total time of visit (in minutes):', '2/24/74 SOS-10 Total Score:', '(2/03/78) TSH-0.90 Activities of Daily Living (ADL) Bathing: Independent', '2/11/2006 CPT Code: 90791: No medical services', "Pt is a 21 year old, single, heterosexual identified, Liechtenstein male. He resides in Rabat, in a rented apartment, with a roommate. He has not graduated from high school nor has he earned his GED. He has been working full time, steadily, since the age of 16. He recently left a job at jacobs engineering group where he worked for about 5 years. He started working at becton dickinson a few weeks ago. Referral to SMH was precipitated by pt seeing his PCP, Dr. Ford, for a physical and sharing concerns re: depression. He was having suicidal thoughts the week of 8/22/83 and noticed that his depression and anxiety symptoms were starting to interfere with work. Per Dr. Ford's recommendation, he contacted PAL for assistance. When screened for substance use, pt was referred to SMH for an intake. Pt was scheduled to be seen last week due to urgency of his depression and SI, but missed the intake appointment due to getting extremely intoxicated the night before. Pt presents today seeking individual therapy and psychiatry services to address longstanding depression.", 'PET Scan (DPSH 5/04/74): 1) Marked hypometabolism involving the bilateral caudate nuclei. This is a non-specific finding, although this raises the question of a multisystem atrophy, specifically MSA-P (striatonigral degeneration), 2) Cortical hypometabolism involving the right inferior frontal gyrus. This is a non-specific finding of uncertain significance, and is not definitively related to a neurodegenerative process, although pathology in this region is not excluded', '7/20/2011 [report_end]', '6/17/95 Total time of visit (in minutes):', '6/10/72 SOS-10 Total Score:', 'nPt denied use to me but endorsed use on 10/16/82 as part of BVH initial visit summary supplement by Nicholas BenjaminOpiates: Yes', '12/15/92 CPT Code: 90801 - Psychiatric Diagnosis Interview', '12/8/97 SOS-10 Total Score:', '4/05/89 Primary Care Doctor:', '12/04/87 SOS-10 Total Score:', '4 (6/20/77)Audit C Score Current:', 'see 4/27/2006 consult note Dr. GuevaraWhat factors in prior treatment were helpful/not helpful:', '07/17/92 CPT Code: 90791: No medical services', '12/22/98 CPT Code: 90801 - Psychiatric Diagnosis Interview', '10/02/96 Age:', '11/05/90 CPT Code: 90792: With medical services', '5/04/77 CPT Code: 90792: With medical services', '2/27/96 Communication with referring physician?: Done']
pandas.Series.str.extract & extractall¶
Extract captures groups in the regex pattern as columns in DataFrame.
# Extract: Grouping done using ()
datetxt_df.str.extract(r'(\d{1,2}[/|-]\d{1,2}[/|-]\d{2,4})')
| 0 | |
|---|---|
| 0 | 03/25/93 |
| 1 | 6/18/85 |
| 2 | 7/8/71 |
| 3 | 9/27/75 |
| 4 | 2/6/96 |
| ... | ... |
| 495 | NaN |
| 496 | NaN |
| 497 | NaN |
| 498 | NaN |
| 499 | NaN |
500 rows × 1 columns
# Extract all
datetxt_df.str.extractall(r'(\d{1,2}[/|-]\d{1,2}[/|-]\d{2,4})')
| 0 | ||
|---|---|---|
| match | ||
| 0 | 0 | 03/25/93 |
| 1 | 0 | 6/18/85 |
| 2 | 0 | 7/8/71 |
| 3 | 0 | 9/27/75 |
| 4 | 0 | 2/6/96 |
| ... | ... | ... |
| 120 | 0 | 12/22/98 |
| 121 | 0 | 10/02/96 |
| 122 | 0 | 11/05/90 |
| 123 | 0 | 5/04/77 |
| 124 | 0 | 2/27/96 |
126 rows × 1 columns
# Further grouping with ()
datetxt_df.str.extractall(r'(\d{1,2})[/|-](\d{1,2})[/|-](\d{2,4})')
| 0 | 1 | 2 | ||
|---|---|---|---|---|
| match | ||||
| 0 | 0 | 03 | 25 | 93 |
| 1 | 0 | 6 | 18 | 85 |
| 2 | 0 | 7 | 8 | 71 |
| 3 | 0 | 9 | 27 | 75 |
| 4 | 0 | 2 | 6 | 96 |
| ... | ... | ... | ... | ... |
| 120 | 0 | 12 | 22 | 98 |
| 121 | 0 | 10 | 02 | 96 |
| 122 | 0 | 11 | 05 | 90 |
| 123 | 0 | 5 | 04 | 77 |
| 124 | 0 | 2 | 27 | 96 |
126 rows × 3 columns
# Grouping can be carried out in a nested fashion!
datetxt_df.str.extractall(r'((\d{1,2})[/|-]((\d{1,2})[/|-](\d{2,4})))')
| 0 | 1 | 2 | 3 | 4 | ||
|---|---|---|---|---|---|---|
| match | ||||||
| 0 | 0 | 03/25/93 | 03 | 25/93 | 25 | 93 |
| 1 | 0 | 6/18/85 | 6 | 18/85 | 18 | 85 |
| 2 | 0 | 7/8/71 | 7 | 8/71 | 8 | 71 |
| 3 | 0 | 9/27/75 | 9 | 27/75 | 27 | 75 |
| 4 | 0 | 2/6/96 | 2 | 6/96 | 6 | 96 |
| ... | ... | ... | ... | ... | ... | ... |
| 120 | 0 | 12/22/98 | 12 | 22/98 | 22 | 98 |
| 121 | 0 | 10/02/96 | 10 | 02/96 | 02 | 96 |
| 122 | 0 | 11/05/90 | 11 | 05/90 | 05 | 90 |
| 123 | 0 | 5/04/77 | 5 | 04/77 | 04 | 77 |
| 124 | 0 | 2/27/96 | 2 | 27/96 | 27 | 96 |
126 rows × 5 columns
# Named groups ?P<NamedGroup>
datetxt_df.str.extract(r'(?P<Date>(?P<Month>\d{1,2})[/|-](?P<Day>\d{1,2})[/|-](?P<Year>\d{2,4}))')
| Date | Month | Day | Year | |
|---|---|---|---|---|
| 0 | 03/25/93 | 03 | 25 | 93 |
| 1 | 6/18/85 | 6 | 18 | 85 |
| 2 | 7/8/71 | 7 | 8 | 71 |
| 3 | 9/27/75 | 9 | 27 | 75 |
| 4 | 2/6/96 | 2 | 6 | 96 |
| ... | ... | ... | ... | ... |
| 495 | NaN | NaN | NaN | NaN |
| 496 | NaN | NaN | NaN | NaN |
| 497 | NaN | NaN | NaN | NaN |
| 498 | NaN | NaN | NaN | NaN |
| 499 | NaN | NaN | NaN | NaN |
500 rows × 4 columns
# It can come handy to add derived columns, e.g. with a .join method to join Dataframes
# Since datetxt_df is a Pandas Series, we first need to convert it into a Dataframe
datetxt_df.to_frame(name="Original_txt")\
.join(datetxt_df.str.extract\
(r'(?P<Date>(?P<Month>\d{1,2})[/|-](?P<Day>\d{1,2})[/|-](?P<Year>\d{2,4}))'))
| Original_txt | Date | Month | Day | Year | |
|---|---|---|---|---|---|
| 0 | 03/25/93 Total time of visit (in minutes): | 03/25/93 | 03 | 25 | 93 |
| 1 | 6/18/85 Primary Care Doctor: | 6/18/85 | 6 | 18 | 85 |
| 2 | sshe plans to move as of 7/8/71 In-Home Servic... | 7/8/71 | 7 | 8 | 71 |
| 3 | 7 on 9/27/75 Audit C Score Current: | 9/27/75 | 9 | 27 | 75 |
| 4 | 2/6/96 sleep studyPain Treatment Pain Level (N... | 2/6/96 | 2 | 6 | 96 |
| ... | ... | ... | ... | ... | ... |
| 495 | 1979 Family Psych History: Family History of S... | NaN | NaN | NaN | NaN |
| 496 | therapist and friend died in ~2006 Parental/Ca... | NaN | NaN | NaN | NaN |
| 497 | 2008 partial thyroidectomy | NaN | NaN | NaN | NaN |
| 498 | sPt describes a history of sexual abuse as a c... | NaN | NaN | NaN | NaN |
| 499 | . In 1980, patient was living in Naples and de... | NaN | NaN | NaN | NaN |
500 rows × 5 columns
Putting a few of the pieces together¶
- Prelude to Ungraded task 2.2
- A bit of data cleaning
- A bit of missing data handling
- Doing so with RegEx
# Let's go back to the CountriesOfTheWorld data.
# We previously loaded it in the CoTW_df DataFrame.
# We see lot's of missing data.
# We had noticed previously that some of the numeric data is stored improperly (as string objects).
CoTW_df.tail(6)
| Country | Region | Population | Area (sq. mi.) | Pop. Density (per sq. mi.) | Coastline (coast/area ratio) | Net migration | Infant mortality (per 1000 births) | GDP ($ per capita) | Literacy (%) | Phones (per 1000) | Arable (%) | Crops (%) | Other (%) | Climate | Birthrate | Deathrate | Agriculture | Industry | Service | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 221 | Wallis and Futuna | OCEANIA | 16025 | 274 | 58,5 | 47,08 | NaN | NaN | 3700.0 | 50,0 | 118,6 | 5 | 25 | 70 | 2 | NaN | NaN | NaN | NaN | NaN |
| 222 | West Bank | NEAR EAST | 2460492 | 5860 | 419,9 | 0,00 | 2,98 | 19,62 | 800.0 | NaN | 145,2 | 16,9 | 18,97 | 64,13 | 3 | 31,67 | 3,92 | 0,09 | 0,28 | 0,63 |
| 223 | Western Sahara | NORTHERN AFRICA | 273008 | 266000 | 1,0 | 0,42 | NaN | NaN | NaN | NaN | NaN | 0,02 | 0 | 99,98 | 1 | NaN | NaN | NaN | NaN | 0,4 |
| 224 | Yemen | NEAR EAST | 21456188 | 527970 | 40,6 | 0,36 | 0 | 61,5 | 800.0 | 50,2 | 37,2 | 2,78 | 0,24 | 96,98 | 1 | 42,89 | 8,3 | 0,135 | 0,472 | 0,393 |
| 225 | Zambia | SUB-SAHARAN AFRICA | 11502010 | 752614 | 15,3 | 0,00 | 0 | 88,29 | 800.0 | 80,6 | 8,2 | 7,08 | 0,03 | 92,9 | 2 | 41 | 19,93 | 0,22 | 0,29 | 0,489 |
| 226 | Zimbabwe | SUB-SAHARAN AFRICA | 12236805 | 390580 | 31,3 | 0,00 | 0 | 67,69 | 1900.0 | 90,7 | 26,8 | 8,32 | 0,34 | 91,34 | 2 | 28,01 | 21,84 | 0,179 | 0,243 | 0,579 |
# Let's check the dtypes to confirm this.
CoTW_df.dtypes
Country object Region object Population int64 Area (sq. mi.) int64 Pop. Density (per sq. mi.) object Coastline (coast/area ratio) object Net migration object Infant mortality (per 1000 births) object GDP ($ per capita) float64 Literacy (%) object Phones (per 1000) object Arable (%) object Crops (%) object Other (%) object Climate object Birthrate object Deathrate object Agriculture object Industry object Service object dtype: object
# Let's convert the Literacy (%) entries into float() type.
# We need to replace the ',' with '.'; following which, we can apply astype(float)
CoTW_df['Literacy (%)']=CoTW_df['Literacy (%)'].str.replace(',', '.').astype(float)
CoTW_df.tail(10)
| Country | Region | Population | Area (sq. mi.) | Pop. Density (per sq. mi.) | Coastline (coast/area ratio) | Net migration | Infant mortality (per 1000 births) | GDP ($ per capita) | Literacy (%) | Phones (per 1000) | Arable (%) | Crops (%) | Other (%) | Climate | Birthrate | Deathrate | Agriculture | Industry | Service | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 217 | Vanuatu | OCEANIA | 208869 | 12200 | 17,1 | 20,72 | 0 | 55,16 | 2900.0 | 53.0 | 32,6 | 2,46 | 7,38 | 90,16 | 2 | 22,72 | 7,82 | 0,26 | 0,12 | 0,62 |
| 218 | Venezuela | LATIN AMER. & CARIB | 25730435 | 912050 | 28,2 | 0,31 | -0,04 | 22,2 | 4800.0 | 93.4 | 140,1 | 2,95 | 0,92 | 96,13 | 2 | 18,71 | 4,92 | 0,04 | 0,419 | 0,541 |
| 219 | Vietnam | ASIA (EX. NEAR EAST) | 84402966 | 329560 | 256,1 | 1,05 | -0,45 | 25,95 | 2500.0 | 90.3 | 187,7 | 19,97 | 5,95 | 74,08 | 2 | 16,86 | 6,22 | 0,209 | 0,41 | 0,381 |
| 220 | Virgin Islands | LATIN AMER. & CARIB | 108605 | 1910 | 56,9 | 9,84 | -8,94 | 8,03 | 17200.0 | NaN | 652,8 | 11,76 | 2,94 | 85,3 | 2 | 13,96 | 6,43 | 0,01 | 0,19 | 0,8 |
| 221 | Wallis and Futuna | OCEANIA | 16025 | 274 | 58,5 | 47,08 | NaN | NaN | 3700.0 | 50.0 | 118,6 | 5 | 25 | 70 | 2 | NaN | NaN | NaN | NaN | NaN |
| 222 | West Bank | NEAR EAST | 2460492 | 5860 | 419,9 | 0,00 | 2,98 | 19,62 | 800.0 | NaN | 145,2 | 16,9 | 18,97 | 64,13 | 3 | 31,67 | 3,92 | 0,09 | 0,28 | 0,63 |
| 223 | Western Sahara | NORTHERN AFRICA | 273008 | 266000 | 1,0 | 0,42 | NaN | NaN | NaN | NaN | NaN | 0,02 | 0 | 99,98 | 1 | NaN | NaN | NaN | NaN | 0,4 |
| 224 | Yemen | NEAR EAST | 21456188 | 527970 | 40,6 | 0,36 | 0 | 61,5 | 800.0 | 50.2 | 37,2 | 2,78 | 0,24 | 96,98 | 1 | 42,89 | 8,3 | 0,135 | 0,472 | 0,393 |
| 225 | Zambia | SUB-SAHARAN AFRICA | 11502010 | 752614 | 15,3 | 0,00 | 0 | 88,29 | 800.0 | 80.6 | 8,2 | 7,08 | 0,03 | 92,9 | 2 | 41 | 19,93 | 0,22 | 0,29 | 0,489 |
| 226 | Zimbabwe | SUB-SAHARAN AFRICA | 12236805 | 390580 | 31,3 | 0,00 | 0 | 67,69 | 1900.0 | 90.7 | 26,8 | 8,32 | 0,34 | 91,34 | 2 | 28,01 | 21,84 | 0,179 | 0,243 | 0,579 |
Datetime object¶
https://docs.python.org/3/library/datetime.html#strftime-strptime-behaviordate1='03-25-93'
date2="3/25/1993"
date3="25 Mar 1993"
print(date1==date3)
False
datetime_object1 = datetime.datetime.strptime(date1,"%m-%d-%y")
print(datetime_object1)
1993-03-25 00:00:00
datetime_object2 = datetime.datetime.strptime(date2,"%m/%d/%Y")
print(datetime_object2)
1993-03-25 00:00:00
datetime_object3 = datetime.datetime.strptime(date3,"%d %b %Y")
print(datetime_object3)
print(datetime_object1==datetime_object3)
1993-03-25 00:00:00 True
Ungraded task 2.2: Clean the CoTW_df Dataframe
Clean the data in the CoTW_df Dataframe as follows:
Create rule based methods (e.g., using regular expressions) to replace the column names as follows:
- Remove everything in brackets, e.g., replace "Coastline (coast/area ratio)" with "Coastline"
- If there are multiple words in the title, then join them with underscores. As such, "Infant mortality (per 1000 births)" would become "Infant_mortality".
Replace all the numeric values which have been stored as strings, into floats.
For all the missing numeric values (NaN), replace them with the Mean value available for the "Region". e.g., Wallis and Futuna is from OCEANIA region, so replace its missing Net_Migration information with the mean Net_Migration as determined for the values available for other countries in OCEANIA.
- Discuss alternatives for dealing with the NaN values. These alternatives may be dependent on type of column and/or the rows involved.
Ungraded task 2.3: Find all the dates in the datetxt_df Series
- Identify the (first instance of) date in each line of the Data Series.
- If only month and year information is available, then assume that the day of the month is 1st.
- If only year information is available, consider the month to be January.
- Create a DataFrame (as shown below) with two columns, the Position of Date in the original Series, and the Date identified, sorted in increasing order of the date.
| Position | Date | |
|---|---|---|
| 0 | 9 | 10/4/1971 |
| 1 | 84 | 18/5/1971 |
| 2 | 2 | 8/7/1971 |
| ... | ... | ... |
| ... | ... | ... |
| 498 | 161 | 19/10/2016 |
| 499 | 413 | 1/11/2016 |
- Discuss: Assuming that you have access to a correct/ideal solution (e.g., the one provided), and you obtain some solution based on your processing, how will you quantitatively compare the quality of result you obtained w.r.to the ideal solution? If you consider several metrics, discuss pros and cons of such various metrics. Implement the corresponding metric and quantify the quality of result you obtain.
- A tentative solution has been provided in an accompanying file named sorted-dates.csv
- Caveat: The solution provided may be faulty. If you find a better solution, please share with me. Please also let me know how you verified inaccuracies of the tentative solution.
That's it folks!